A tandem repeat in DNA is two or more contiguous, approximate copies of a pattern of nucleotides. Tandem repeats have been shown to cause human disease, may play a variety of regulatory and evolutionary roles and are important laboratory and analytic tools. Extensive knowledge about pattern size, copy number, mutational history, etc. for tandem repeats has been limited by the inability to easily detect them in genomic sequence data. In this paper, we present a new algorithm for finding tandem repeats which works without the need to specify either the pattern or pattern size. We model tandem repeats by percent identity and frequency of indels between adjacent pattern copies and use statistically based recognition criteria. We demonstrate the algorithm’s speed and its ability to detect tandem repeats that have undergone extensive mutational change by analyzing four sequences: the human frataxin gene, the human beta T cellreceptor locus sequence and two yeast chromosomes.
The following programs to analyze DNA sequences offered:
The program is destined to search for non-perfect repeats of various types in a nucleotide sequences.
Types of repeats definition
Depending on type of correspondence we consider the following types of repeats:
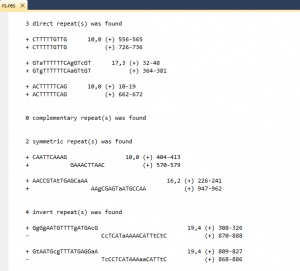
Direct repeat – two (l,k) segments are located in the same orientation in a single DNA strand;
Symmetrical – two (l,k) segments are located in opposite orienation in a single DNA strand;.
Direct complementary repeats – two segments are located in the same orientation in different DNA strands;.
Inverse repeats – two segments are located in oppsite orientation in the different DNA strings
Program features
The program performs the repeats search by group clusters (l,k). After the search the optimization procedure follows consisting of the following steps:
the concatenation of the groups (l,k) in one set is performed. During that:
the repeats flanks are extended if there are coinciding nucleotides ;
All abundant repeats are deleted

The program is destined to visualise the results of repeats search by ReSearch program, and is included in OligoRep program.
Vizualised information
various types of repeats (direct, symmetrical, direct complementary, inversed) are plotted by various colors.
Each repeat is plotted on a separate double-stranded string
by double clicking on a particular repeat additional information such as:
copy length
mismatch number
position in the target DNA string
nucleotide content of the copy coinciding letters marked
is visulaised;
Nucleotide positions are located in the upper area of image on the band, nucleotides marked by:
A – Adenine(yellow)
T – Thymine(brown)
G – Guanine(black)
C – Cytosine(white)
How it works Pressing “Visualisation” button the reuslts of repeats search in target sequence are visualised. By mouse double -clicking the particular repeat structure and content is visualised separately.
Control elements
Clear
Clear the image area
Max +
Min –
Change the scale
Double click
Repeat structure and content vizualisation
Oligonucleotids repeats finder:
http://lcg.nsu.ru/en/oligonucleotids-repeats-finder/
Authors: V.V. Bazin, P.S. Kosarev, V.N. Babenko
